Abstract
Principal component analysis (PCA) is a dimensionality reduction method in data analysis that involves diagonalizing the covariance matrix of the dataset. Recently, quantum algorithms have been formulated for PCA based on diagonalizing a density matrix. These algorithms assume that the covariance matrix can be encoded in a density matrix, but a concrete protocol for this encoding has been lacking. Our work aims to address this gap. Assuming amplitude encoding of the data, with the data given by the ensemble , then one can easily prepare the ensemble average density matrix . We first show that is precisely the covariance matrix whenever the dataset is centered. For quantum datasets, we exploit global phase symmetry to argue that there always exists a centered dataset consistent with , and hence can always be interpreted as a covariance matrix. This provides a simple means for preparing the covariance matrix for arbitrary quantum datasets or centered classical datasets. For uncentered classical datasets, our method is so-called “PCA without centering,” which we interpret as PCA on a symmetrized dataset. We argue that this closely corresponds to standard PCA, and we derive equations and inequalities that bound the deviation of the spectrum obtained with our method from that of standard PCA. We numerically illustrate our method for the Modified National Institute of Standards and Technology (MNIST) handwritten digit dataset. We also argue that PCA on quantum datasets is natural and meaningful, and we numerically implement our method for molecular ground-state datasets.
1 More- Received 14 April 2022
- Revised 5 July 2022
- Accepted 16 August 2022
DOI:https://doi.org/10.1103/PRXQuantum.3.030334
Published by the American Physical Society under the terms of the Creative Commons Attribution 4.0 International license. Further distribution of this work must maintain attribution to the author(s) and the published article's title, journal citation, and DOI.
Published by the American Physical Society
Physics Subject Headings (PhySH)
Popular Summary
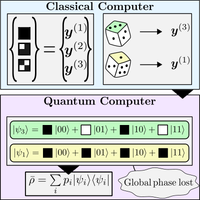
Principal component analysis (PCA) is an essential part of machine learning and data analysis. It is used to reduce the dimensionality in a dataset while minimizing information loss. Implementing PCA on a quantum device is of great interest to the quantum computing community. However, an essential step in quantum PCA was missing: how to prepare the covariance matrix on a quantum computer.
Our main result proves that the ensemble average density matrix, which is extremely easy to prepare on a quantum computer, happens to correspond to the covariance matrix for quantum datasets. This is a timely result, since it was recently shown that quantum computers could provide an exponential speedup for PCA on quantum datasets. Therefore, our work provides the missing link in the quest for demonstrating exponential quantum speedup with PCA, opening the door for near-term implementations on quantum hardware.
In addition, we provide several conceptual insights. (1) We show that PCA on molecular ground states can lead to dramatic data compression. (2) We show that our method is PCA without centering for classical datasets, and we illustrate this for the famous MNIST handwritten digit dataset. (3) We offer a new interpretation of PCA without centering as being PCA on a symmetrized dataset. (4) We rigorously prove that PCA without centering yields a spectrum very similar to standard PCA.